BECK Lab Research
As one of the most abundant proteins in the cell, actin participates in many important cellular processes including cell motility, maintenance of cell shape, cell division, and muscle contraction. Coordination of these diverse cellular processes therefore requires responsive regulation of actin. Currently over 150 actin-binding proteins have been identified which influence localization, polymerization dynamics, crosslinking, and organization of actin. Cytoskeletal actin is important for normal cellular function, however it can also be subverted in cancer and contributes to changes in cell motility and invasiveness.
Palladin is a recently identified protein from a novel family of actin binding proteins that has emerged as a key player in organizing actin arrays within migrating cells. Overexpression of palladin has been linked to highly metastatic cancers such as breast and pancreatic. Palladin family members utilize immunoglobulin-like (Ig) domains to bind and crosslink actin, highlighting a new role for Ig domains. Our preliminary results suggest yet another novel role for palladin that may include both promoting actin polymerization and/or modifying the structure of actin filaments.
Recently, mutations in the muscle protein myopalladin have been linked to the pathogenesis of cardiomyopathy. While myopalladin is thought to play a role in regulating the organization of sarcomere structure within cardiac and skeletal muscles, insufficient knowledge of myopalladin’s interactions with other key molecules has hampered development of new therapies for cardiomyopathies. Mutations in myopalladin cause diverse cardiomyopathic phenotypes that are poorly understood; therefore, our goal is to understand the relationship between structural and biochemical alterations in cellular and molecular mechanisms underlying sarcomere organization, regulation and function
Current research interests in the Beck group are to investigate the mechanisms by which palladin and myopalladin, through both direct and indirect molecular mechanisms, affects actin dynamics. Specifically, we will use biochemical and cell biological approaches, coupled with protein structural studies to elucidate the role and mechanisms for how the novel, actin crosslinking protein palladin and myopalladin influences the actin cytoskeleton. We intend to elucidate in detail the biochemical mechanisms of actin regulation and organization using kinetic assays of actin polymerization coupled with studies of protein-protein interactions via NMR.
TIRF Microscopy allows us to visualize individual actin filaments grow in real time. So we can compare the amount of filament growth and the resulting structures of the actin filaments in the presence and absence of palladin.
Actin: Important Roles in Motility and Metastasis
Fluorescence microscopy of palladin and actin localization in cells expressing WT (top) or actin binding mutant (bottom).
Palladin: A Novel Actin Binding Protein
Multidisciplinary and Collaborative Approach
New results show us how palladin effects actin filament growth and structure.
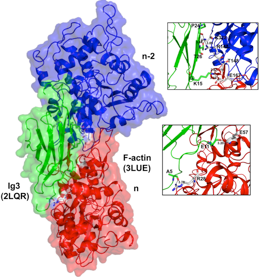
NMR Structure of Ig3 domain of palladin docked onto actin.
Myopalladin: Role in Cardiomyopathy
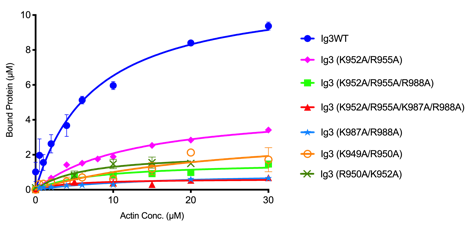
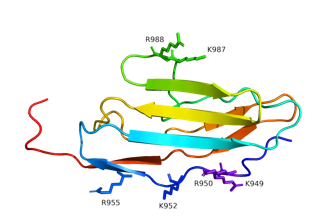
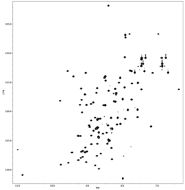
Actin binding curves (below, left) reveal that all of the surface exposed lysine and arginine residues (left) are involved in binding interaction in myopalladin.
TIRF microscopy time lapse imaging reveals growth of actin filaments in the presence of palladin results in more branched structures than actin alone.
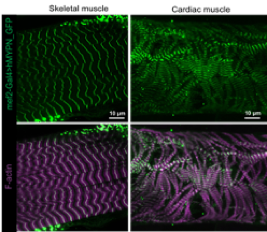
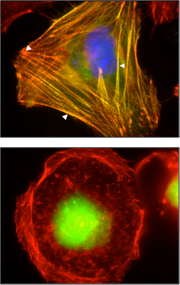
Expression of human wildtype myopalladin (hMYPN-GFP) in Drosophila third instar larval skeletal (left panels) and cardiac muscle (right panels). Immunostaining with F-actin is used to visualize muscle pattern. Data collected in collaboration with Dr. Erika Geisbrecht at KSU.
15N-HSQC NMR spectrum of myopalladin Ig3 domain collected at OSU’s new 800 MHz NMR facility.
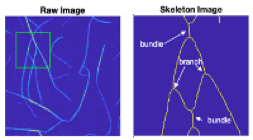
Raw, false-colored image of actin polymerization in the presence of palladin Ig3. The green box indicates the field of view that is expanded on for closer analysis (top, right). Branches and bundles are indicated after image processing and skeletonizing the actin fibers. Image analysis carried out in Biophotonics lab in collaboration with Dr. Vince Rossi at Washburn University.